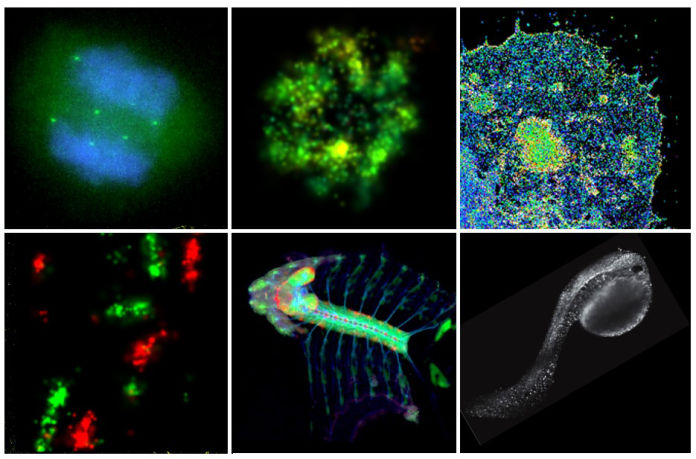
Watch The Inner Life Of A Cell
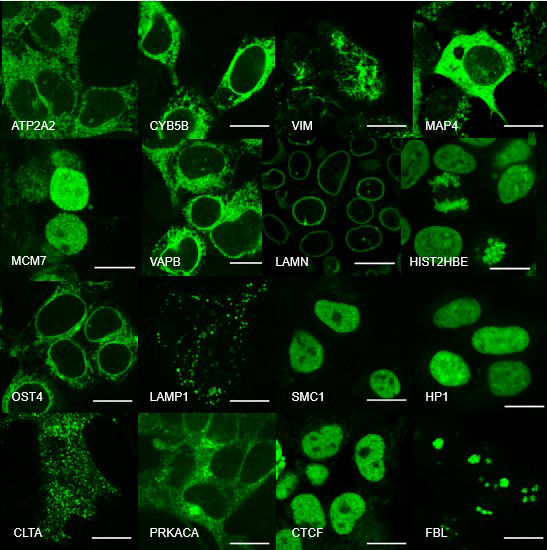
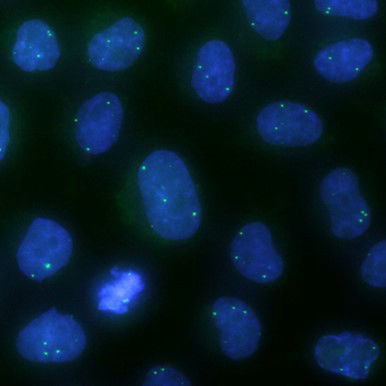
Cellular processes are orchestrated by a large number of biomolecules in a spatially and temporally coordinated manner within a tiny volume. To uncover the underlying organizational principles and their functional relevance, we take microscopy visualization as the primary approach to systematically map the spatial localization, temporal dynamics and activity profiles of proteins and nucleic acids.