High-throughput light-sheet microscopy and subcellular protein atlas
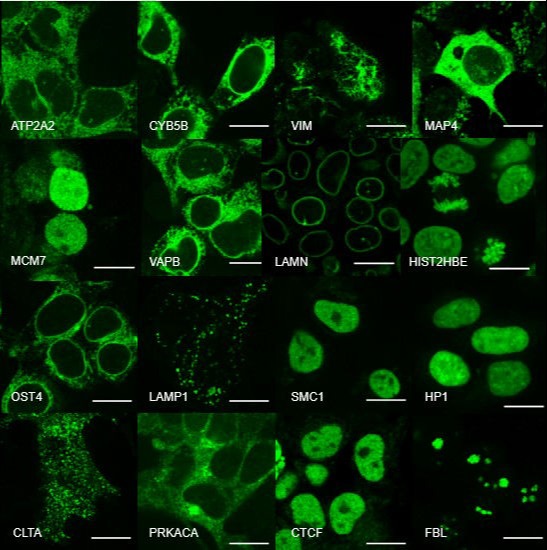
For our ultimate goal of painting a proteome-wide map of endogenous proteins inside human cells, we are developing three techniques: (1) new split-protein-based fluorescent probes that enable large-scale labeling of endogenous proteins in human cell lines via gene editing, (2) new light-sheet microscopes for high-resolution 3D live cell imaging, and (3) deep-learning methods for efficient processing and quantitative interpretation of imaging data. Combing these three technical advances, we are systematically analyzing the dynamic re-organization of various nuclear and cytoplasmic structures throughout the cell cycle.
• Yang et al., Nat. Methods (2019)
• Feng et al., Nat. Commun. (2017)
• Kamiyama & Sekine et al., Nat Commun. (2016)
• Leonetti & Sekine et al., PNAS (2016)