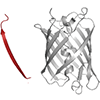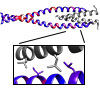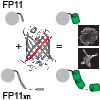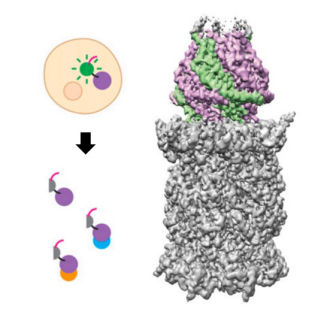
Structural insights into human PA28-20S proteasome enabled by efficient tagging and purification of endogenous proteins
J. Zhao*, S. Makhija*, C. Zhou, H. Zhang, Y. Wang, M. Muralidharan, B. Huang, Y. Cheng
PNAS 119, e2207200119 (2022) [link] [Preprint: bioRxiv/2021.06.25.449985]